Our immune system has a complicated and unique mechanism of defense against viruses, bacteria and other pathogens entering our bodies. It might also detect differences in human tissue resulting in an attack of transplanted kidneys resulting in graft failure. Hence, the donor and recipient immune systems are checked for compatibility using genetic matching prior the transplant. This approach is limited by the complex nature of the human immune system. Therefore, more recent matching approaches focus on the structure of proteins considering their individual function. This method allows a more precise prediction of risk factors for immune system incompatibility leading to improved graft survival.
Thanks to the contributing experts from our partners for their input for this article: Matthias Niemann (PIRCHE), Aadil Rasheed (HPI), Dr. Matthieu-P. Schapranow (HPI). Photo by Israel Palacio.
Why do I encounter graft failure?
In our recent article, we introduced kidney transplantation and its challenges. A major risk factor for premature graft failure is the patient’s immune system detecting the transplanted kidney as a foreign tissue. As a result, it will start the development of specific antibodies targeting the new organ resulting in a graft failure over time.
Human Leukocyte Antigens (HLAs) are specific proteins on the surface of most human cells. They are used by the immune system to distinguishing the patient’s own cells from foreigners. The human immune system consists of multiple actors, such as lymphocytes also known as white blood cells. Specific lymphocytes are produced in the bone marrow (B cells) and the Thymus (T cells). B cells are capable to digest foreign antigens and produce antibodies. T cells detect antigens on the cell surface and can release cytotoxins to fight such invaders. Once the B cells recognized cells of the donor organ as invaders, they start to produce antibodies. They bind to HLA proteins located on the graft’s cell surface.
From DNA to HLAs
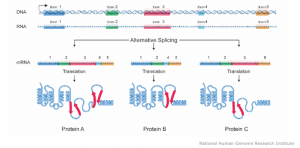
Fig. 1: DNA is transcribed to RNA, which is translated by ribosomes to amino-acid chains forming proteins. Taken from National Humane Genome Res. Inst.
Proteins form most of the antigens, also the HLAs. The DNA stores the blueprint for the corresponding proteins. Once the respective part of the DNA is read, it is transcribed into RNA, which is then translated into sequences of amino acids. Ultimately, these amino acid sequences fold into their tertiary three-dimensional structure forming a fully functional HLA protein. Fig. 1 depicts an additional type of variation introduced by so-called alternative splicing. It refers to the observation that even the same DNA might result in different proteins and properties based on how it is translated by our ribosomes.
How to identify the best donor-recipient match to reduce the risk of graft failure?
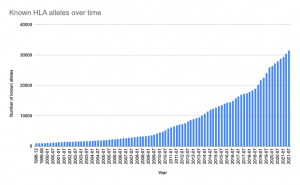
Fig. 2: The number of known HLA alleles is huge and since the development of high-throughput sequencing techniques, we find more and more alleles. Based on https://www.ebi.ac.uk/ipd/imgt/hla/stats.html
Today, we know that the risk for graft failure due to an immune response in the recipient can be reduced. Therefore, a matching of donors and recipients is conducted in such a way that the recipient’s immune system has as much as compatible HLAs in common with the donor. As a results, the number of potential targets to develop antibodies is lower as well as the risk for developing an immune response against the new organ resulting in graft failure.
Traditionally, a set of just six specific locations of the HLA-encoding gene were considered for assessing the compatibility between donor and recipient. With the spread of high-throughput sequencing technologies in the recent years, we learnt about more and more allele combinations on HLA-encoding genes. Fig. 2 shows a total of more than 30k+ HLA alleles documented by 2021.
On the one hand, the HLA gene is highly polymorphic to adapt to all kinds of diseases making it very hard to find a 100% match between donors and recipients. On the other hand, not every mismatch on the genetic level has an impact on the three-dimensional protein structure exemplarily depicted in Fig. 3.
Next generation of organ matching: Epitope matching
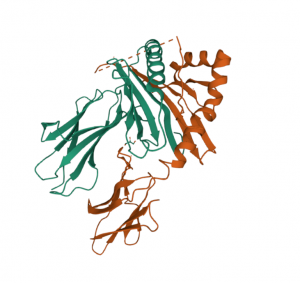
Fig. 3: Three-dimensional structure of an HLA DQ heterodimer taken from http://doi.org/10.2210/pdb4D8P/pdb
The HLA-based matching for organ transplantation, also known as serological or “classical” matching of HLAs, is only able to identify whether two HLA proteins will be identical of not. However, it cannot assess the broad spectrum of small changes within the protein structure and how probable the recipient’s immune system will identify such mismatches between two HLA proteins.
In contrast, epitope matching focuses on the actual structure of the HLA proteins and its interaction capabilities. The complexity and heterogeneity of the genes relevant for HLAs require advanced bioinformatic simulations. For example, Fig. 3 depicts the complex three-dimensional structure of the HLA DQ heterodimer as result of a simulation. However, there is evidence that current epitope matching algorithms have superior performance in predicting immune responses [1, 2, 3].
Epitope matching: B- vs. T-cell epitope
There are two major streams in predicting HLA epitopes, dedicated to the major pathways of allorecognition:
- B-cell epitopes: B cell epitopes are small patches on the surface of intact HLA molecules. A deviation of these patches between the recipient and donor’s HLA is interpreted as epitope mismatch. These mismatches are potential targets for HLA antibodies, which are produced by the corresponding B cells.
- T-cell epitopes: T-cell epitopes are linear peptides derived from digested donor HLA proteins presented to T cells. Upon detection of unknown peptides, T cells become activated promoting a cascade of immune-regulatory response.
Meanwhile, several algorithms for epitope matching exist. For example, HLA Matchmaker for B-cell epitopes and PIRCHE for T-cell epitopes. The different pathways of allorecognition also showed a statistical independence, which suggests a combined matching model may perform better than the individual algorithms alone.
The NephroCAGE consortium
The German Canadian consortium NephroCAGE combines medical and scientific-technical expertise from Germany and Canada. We jointly create a learning clinical system that tests the added value of the latest artificial intelligence (AI) techniques on the concrete clinical example of kidney transplantation. Therefore, our NephroCAGE partners create a learning AI system for matching organ donors and recipients even more precisely in order to reduce risks in kidney transplants and to prevent graft failure or even its loss. One component will be a prototype of a combined epitope matching algorithm. It will be compared to individual algorithms and “classic” HLA matching. The transatlantic partnership allows us to understand population-specific effects in epitope matching.




No responses yet